PASIEKA/Science Photo Library via Getty Images
William Wright, University of California, San Diego and Takaki Komiyama, University of California, San Diego
Every day, people are constantly learning and forming new memories. When you pick up a new hobby, try a recipe a friend recommended or read the latest world news, your brain stores many of these memories for years or decades.
But how does your brain achieve this incredible feat?
In our newly published research in the journal Science, we have identified some of the “rules” the brain uses to learn.
Learning in the brain
The human brain is made up of billions of nerve cells. These neurons conduct electrical pulses that carry information, much like how computers use binary code to carry data.
These electrical pulses are communicated with other neurons through connections between them called synapses. Individual neurons have branching extensions known as dendrites that can receive thousands of electrical inputs from other cells. Dendrites transmit these inputs to the main body of the neuron, where it then integrates all these signals to generate its own electrical pulses.
It is the collective activity of these electrical pulses across specific groups of neurons that form the representations of different information and experiences within the brain.
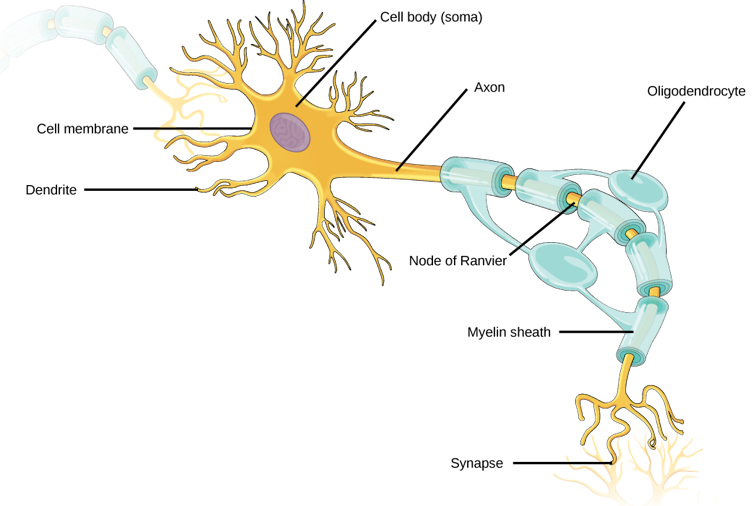
OpenStax, CC BY-SA
For decades, neuroscientists have thought that the brain learns by changing how neurons are connected to one another. As new information and experiences alter how neurons communicate with each other and change their collective activity patterns, some synaptic connections are made stronger while others are made weaker. This process of synaptic plasticity is what produces representations of new information and experiences within your brain.
In order for your brain to produce the correct representations during learning, however, the right synaptic connections must undergo the right changes at the right time. The “rules” that your brain uses to select which synapses to change during learning – what neuroscientists call the credit assignment problem – have remained largely unclear.
Defining the rules
We decided to monitor the activity of individual synaptic connections within the brain during learning to see whether we could identify activity patterns that determine which connections would get stronger or weaker.
To do this, we genetically encoded biosensors in the neurons of mice that would light up in response to synaptic and neural activity. We monitored this activity in real time as the mice learned a task that involved pressing a lever to a certain position after a sound cue in order to receive water.
We were surprised to find that the synapses on a neuron don’t all follow the same rule. For example, scientists have often thought that neurons follow what are called Hebbian rules, where neurons that consistently fire together, wire together. Instead, we saw that synapses on different locations of dendrites of the same neuron followed different rules to determine whether connections got stronger or weaker. Some synapses adhered to the traditional Hebbian rule where neurons that consistently fire together strengthen their connections. Other synapses did something different and completely independent of the neuron’s activity.
Our findings suggest that neurons, by simultaneously using two different sets of rules for learning across different groups of synapses, rather than a single uniform rule, can more precisely tune the different types of inputs they receive to appropriately represent new information in the brain.
In other words, by following different rules in the process of learning, neurons can multitask and perform multiple functions in parallel.
Future applications
This discovery provides a clearer understanding of how the connections between neurons change during learning. Given that most brain disorders, including degenerative and psychiatric conditions, involve some form of malfunctioning synapses, this has potentially important implications for human health and society.
For example, depression may develop from an excessive weakening of the synaptic connections within certain areas of the brain that make it harder to experience pleasure. By understanding how synaptic plasticity normally operates, scientists may be able to better understand what goes wrong in depression and then develop therapies to more effectively treat it.
William J. Giardino/Luis de Lecea Lab/Stanford University via NIH/Flickr, CC BY-NC
These findings may also have implications for artificial intelligence. The artificial neural networks underlying AI have largely been inspired by how the brain works. However, the learning rules researchers use to update the connections within the networks and train the models are usually uniform and also not biologically plausible. Our research may provide insights into how to develop more biologically realistic AI models that are more efficient, have better performance, or both.
There is still a long way to go before we can use this information to develop new therapies for human brain disorders. While we found that synaptic connections on different groups of dendrites use different learning rules, we don’t know exactly why or how. In addition, while the ability of neurons to simultaneously use multiple learning methods increases their capacity to encode information, what other properties this may give them isn’t yet clear.
Future research will hopefully answer these questions and further our understanding of how the brain learns.![]()
William Wright, Postdoctoral Scholar in Neurobiology, University of California, San Diego and Takaki Komiyama, Professor of Neurobiology, University of California, San Diego
This article is republished from The Conversation under a Creative Commons license. Read the original article.